
Autophagy machinery -
In addition to its bulk degradation property, autophagy also partakes in the clearance of specific substrates. This selective autophagy mainly depends on cargo receptors such as neighbor of BRCA1 gene 1 NBR1 and p62, which can bind to ubiquitin-tagged substrates.
These cargo receptors can also bind to LC3 via a LC3-interacting region LIR motif, which therefore targets them to autophagosomes Bjørkøy et al.
Phagophores require lipids to mature into autophagosomes. After more than 50 years of investigations, the origin of the autophagosomal membranes is still a critical question.
Originally, the endoplasmic reticulum ER was proposed to be the primary source of these membranes. Early electron microscopy studies identified a close relationship between the ER and autophagic structures, suggesting that autophagosomal membranes are mainly delivered from the ER Novikoff and Shin, ; Hayashi-Nishino et al.
Consistent with this idea, Axe et al. These subdomains then constitute a platform for accumulation of autophagosomal proteins, expansion of autophagosomal membranes and emergence of fully formed autophagosomes. Subsequent three-dimensional tomography studies Hayashi-Nishino et al.
The associated ER and isolation membranes are interconnected by a narrow membrane extension from the isolation membrane. Recent studies found evidence that apart from the ER, numerous other membrane sources are involved in the formation of autophagosomes, including mitochondria, the Golgi, recycling endosomes and endocytic vesicles budding from the plasma membrane.
Hailey et al. They found that the early autophagosomal maker, ATG5, transiently localizes to puncta on mitochondria, followed by the late autophagosomal marker LC3.
This study further showed that cell starvation drives the delivery of lipid components from the mitochondrial outer membrane to newly formed autophagosomes. It has recently been reported that the Golgi may also contribute to the formation of autophagosomes.
Following starvation, activation of the class-III PI3K complex promotes re-localization of COPII adaptors from the ER exit sites to the ER-Golgi intermediate compartment ERGIC. The process leads to the generation of ERGIC-derived COPII vesicles which becomes LC3-positive and contribute to autophagosome biogenesis Ge et al.
Recent reports demonstrated that recycling endosomes, through the formation of tubular structures accumulating autophagy proteins, also supply membrane for autophagosome biogenesis.
In a siRNA-mediated screen, Knaevelsrud et al. identified the PX domain-containing protein, SNX18, as a positive regulator of autophagy Knævelsrud et al.
The membrane binding and tubulation activities of SNX18, as well as its direct interaction with LC3, allow the formation of LC3-ATG16L1-positive tubules emanating for recycling endosomes that provide membrane input to forming autophagosomes.
This study is in line with other findings Longatti et al. Together, these data indicate that the recycling compartment is not solely responsible for recycling of plasma membrane receptors but also serves as a sorting station for controlled delivery of membrane for autophagosome biogenesis.
Work from Rubinsztein's lab identified endocytic vesicles, trafficking to recycling endosomes, as an important source of membrane for autophagosome biogenesis. Endocytic vesicles can form from regions of the plasma membrane through different mechanisms, i.
Accumulation of ATG16L1 at clathrin-coated endocytic structures, through an interaction between ATG16L1 and the clathrin adaptor AP2, and vesiculation of ATG16L1-positive precursors have been found to contribute to autophagosome formation. Inhibition of clathrin-mediated endocytosis, using siRNAs targeting the clathrin heavy-chain or the clathrin adaptor AP2, causes defective autophagosome biogenesis, which is associated with impaired uptake of plasma membrane into pre-autophagosomal vesicles Ravikumar et al.
These ATG16L1-positive vesicles then undergo SNARE-mediated homotypic fusion, generating tubulovesicular structures that increase in size, enabling the acquisition of LC3 protein Moreau et al. Surprisingly, ATG16L1 and ATG9 proteins have been found to localize to distinct clathrin-coated vesicles and to traffic through different routes inside the cell.
Although both ATG9 and ATG16L1 proteins end up in recycling endosomes, ATG9 is trafficked via EEA1-positive early endosomes, whereas ATG16L1 has minimal residence in early endosomes Puri et al.
The SNARE protein named VAMP3, which co-traffics with ATG9, seems to be critical for the coalescence of ATG16L1 and ATG9 vesicles in recycling endosomes Puri et al. The impact of this coalescence on the formation of tubules emanating from recycling endosomes, driven by SNX18, deserves further investigations.
To this day, very few GPCRs have been shown to directly affect autophagic activity. These mainly include nutrient sensing receptors that increase anabolic processes via stimulation of the mTOR kinase, a well-known autophagy repressor Jung et al.
It has been suggested that this GPCR may impact autophagic activity through mTOR stimulation. Reducing T1R3 levels in HeLa cells is sufficient to impair mTOR activity and activate autophagy Wauson et al. Angiotensin receptors have also been found to modulate autophagic activity in cardiomyocytes Porrello et al.
We recently found that chemotactic GPCRs CXCR4 and the urotensin II receptor UT also reduce autophagic activity by inhibiting autophagosome biogenesis Coly et al. Unlike the studies cited above, these anti-autophagic effects do not seem to be relayed by mTOR modulation, but rather by inhibiting ATG16L1 recruitment to pre-autophagic vesicles budding from the plasma membrane.
While Ravikumar et al. Bjørkøy et al. We demonstrated that activation of CXCR4 or UT reduces the pool of ATG5 protein located at the plasma membrane, thereby reducing the recruitment of ATG16L1.
Accordingly, overexpression of recombinant ATG5 totally abrogates the anti-autophagic activities of CXCR4 and UT, and siRNA-mediated knockdown of ATG5 mimics the inhibitory effects of these GPCRs on the formation of pre-autophagic endosomes.
What is the exact role of ATG5 in mediating the formation of pre-autophagic endosomes? We can speculate that ATG5's membrane binding activity Romanov et al. Alternatively, since ATG5 can co-immunoprecipitate from cell lysates with ATG16L1 and clathrin, and since the N-terminus region of ATG16L1 allows both AP2-clathrin co-immunoprecipitation Ravikumar et al.
Calpains are a ubiquitously expressed family of cysteine proteases that mediate cleavage of specific substrates. Calpains have thus been found to be involved in a number of processes such as development, cell death, and motility Goll et al.
Modulating cell migration is one of the better known roles of these proteases. Studies conducted in neutrophils have shown that calpain inhibition increases random migration, but decreases GPCR-induced directional migration upon exposure to a gradient of interleukin 8 Lokuta et al.
In neurons, calpain activity was also shown to be essential for SDF1-induced actin reorganization and directional migration Lysko et al. These results are in line with work highlighting the role of the calpain 2 isoform during lamellipodium formation.
Calpain 2 controls the formation of cell protrusions by cleaving cortactin, a key modulator of actin filament branching at the cell front.
Expression of a calpain-resistant form of cortactin reduces the migration of fibroblasts by increasing the number of transient and inefficient cell protrusions Perrin et al. Calpains also play an important role in the dynamics of adhesion formation and disassembly.
By modifying the cytoplasmic tail of β-integrins, calpains seem to be essential for the formation of integrin clusters at an early stage of adhesion complex assembly Bialkowska et al.
Once cleaved, talin can bind to β-integrin tails, therefore constituting the first link between integrins and actin filaments Yan et al. They contribute to adhesion turnover by destabilizing the structural integrity of the complex. Several proteins such as paxillin, vinculin and talin are in fact targeted by calpains during this stage Carragher et al.
Inhibiting calpains with either calpastatin or pharmacological means significantly slows adhesion turnover Bhatt et al. Similar results can be obtained following calpain 2 knockdown, which results in large, long lasting adhesion complexes that inhibit cell detachment and therefore impair cell migration Franco S.
Despite the many roles of calpains during cell migration, their regulation by chemotactic GPCRs remains unclear. However, previous work revealed that calpain 2 is recruited at the plasma membrane and activated following its phosphorylation by ERK and dephosphorylation on a protein kinase A PKA site Glading et al.
Interestingly, as mentioned earlier, the pro-migratory properties of many chemotactic GPCRs are relayed by G i coupling, which has the ability to activate ERK, through βγ subunits, and to inhibit PKA, through the α i subunit Goldsmith and Dhanasekaran, ; Cotton and Claing, We can therefore speculate that the simultaneous induction of these signaling pathways by chemotactic GPCRs may be determinant for the activation of calpain 2 at the plasma membrane and regulation of adhesion dynamics.
A growing amount of data suggests that calpains are major inhibitors of the autophagy machinery. SiRNA-mediated knockdown of calpain 1 is sufficient to induce autophagy under nutrient rich conditions, correlated with increased levels of LC3-II and ATG5-ATG12 complex Xia et al.
Using a cell-free system, Yousefi et al. demonstrated that ATG5 can be cleaved by both calpain 1 and calpain 2 Yousefi et al.
Cleavage of ATG5 then generates a 24 kDa N-terminal product that can translocate to the mitochondria and enhance susceptibility toward apoptotic stimuli Yousefi et al. In vitro experiments also identified ATG3, ATG4, ATG7, ATG9, ATG10, ATG12, and Beclin1 as direct calpain substrates Norman et al.
It should be noted that calpains may also exert their anti-autophagic properties by targeting non-ATG proteins. The clathrin adaptors AP2 and PICALM, which are critical for the formation of pre-autophagosomal vesicles from the plasma membrane, have been described as calpain substrates Kim and Kim, ; Rudinskiy et al.
Does calpain-dependent repression of autophagy then constitute a critical event for chemotaxis? In favor of this hypothesis, we found that the anti-autophagic and pro-migratory properties of two chemotactic GPCR, CXCR4, and UT, were abrogated by pharmacological inhibition or siRNA knockdown of calpains Coly et al.
We further demonstrated that calpain activation, induced by CXCR4 or UT, reduces the pool of ATG5 at the plasma membrane and inhibits the recruitment of ATG16L1 protein to endocytic vesicles, thereby limiting the formation of pre-autophagosomal precursors required for the expansion of the phagophore and formation of mature autophagosomes.
In addition to reversing the anti-autophagic effects of chemotactic GPCRs, calpain inhibition or ATG5 overexpression is also sufficient to block their pro-migratory properties, as both these approaches reduce the cells' migration rate, as well as the number of adhesions per cell Coly et al.
Despite early reports pointing to ATG5 as a calpain target, our attempts at demonstrating its direct cleavage following CXCR4 or UT activation were unsuccessful. One hypothesis is that only a minor, plasma membrane-associated fraction of ATG5 is cleaved by calpains.
The cleaved products may also be highly unstable, thereby hindering their detection. Alternatively, the anti-autophagic action of calpains following GPCR activation could depend on the cleavage of the adaptor proteins AP2 and PICALM, or on the cleavage of ATG7, which is essential for conjugation of ATG5—ATG12 Mizushima et al.
Since the recruitment of calpains at the plasma membrane constitutes an early event during chemotaxis Franco and Huttenlocher, , it can be anticipated that GPCR-induced inhibition of autophagy may tightly control early steps of cell polarization.
During GPCR-induced chemotactic migration, efficient expansion of the lamellipodium requires addition of extra membrane at the leading edge, through polarized, microtubule-dependent exocytosis Bretscher and Aguado-Velasco, ; Pierini et al. Work from Veale et al. identified VAMP3-positive recycling endosomes as an important source of internal membrane that is incorporated at the leading edge during macrophage migration Veale et al.
Along with the incorporation of extra membrane, this mechanism also allows the recycling of cell adhesion components at the leading edge, including integrins Veale et al.
Since recycling endosomes, through the SNXdependent formation of tubules, supply membrane for phagophore expansion, it is conceivable that this compartment may constitute a sorting station that deliver phospholipids in a competitive manner, for either lamellipodium expansion or autophagosome synthesis.
The dynamic increase in plasma membrane surface triggered by chemotactic GPCRs may then directly impact the pool of phospholipids available for autophagic activity. How could activation of GPCRs, located at the cell surface, affect the trafficking of membrane from recycling endosomes? Chemotactic GPCRs CXCR4 and UT alter, through the activation of calpains, the recruitment of ATG16L1 in pre-autophagosomal vesicles budding from the plasma membrane Coly et al.
This may reduce the pool of ATG16L1 targeted to the recycling compartment and limit the coalescence of ATG16L1 and ATG9 vesicles. Inhibition of ATG16L1 and ATG9 coalescence would then favor the delivery of VAMP3-positive vesicles at the cell front, at the expense of the phagophore Figure 1.
Figure 1. Chemotactic GPCR-mediated autophagy inhibition: potential role in chemotactic migration. A Under basal conditions, ATG5-ATG16L1-positive pre-autophagic endosomes bud from the plasma membrane and are directed to the recycling endosome compartment.
From there, SNXdependent tubules target vesicles containing ATG5-ATG16L1 and LC3 to the expanding phagophore. B Upon activation by chemoattractant stimuli, chemotactic GPCRs locally inhibit the formation of pre-autophagic endosomes. Exocytosis allows integrins to be recycled to nascent adhesions, while phospholipids are incorporated into the lamellipodium and contribute to its expansion.
Autophagy inhibition at the leading edge may also locally protect proteins involved in actin remodeling and adhesion assembly, which would otherwise be sequestered and degraded. Autophagy could remain active at distance from chemotactic GPCRs in order to participate in the disassembly of large focal adhesions.
Among the hallmarks of cell migration, the formation of adhesion complexes at the cell's leading edge is among the most notable. Adhesions are critical in generating the traction required for the cell's forward movement. Several data have demonstrated autophagic degradation of key proteins involved in the initiation and the maturation of adhesion complexes, indicating that autophagy can regulate adhesion dynamics.
The Src kinase, which is involved in adhesion signaling, was shown to co-immunoprecipitate with LC3 and to be degraded by autophagy Sandilands et al. In fibroblasts, β1 integrin-containing vesicles co-localize with LC3-stained autophagic structures. Inhibition of autophagy by ATG5 or ATG3 knockdown is able to slow β1 integrin degradation and to promote it's recycling to the plasma membrane Tuloup-Minguez et al.
Kenific et al. Shiraha et al. Furthermore, paxillin was shown to have its own LIR domain, which is also involved in its autophagic degradation Sharifi et al. In agreement with a role of autophagy in adhesion disassembly, global inhibition of autophagosome biogenesis, using knockdown strategies against ATG proteins, results in the accumulation of large and unproductive adhesions at the entire cell periphery that reduce cell migration Kenific et al.
Also these migration studies could appear to conflict with our report, Coly et al. Chemotactic GPCRs are known to induce actin polymerization at the cell's leading edge to allow the lamellipodium to protrude toward the chemoattractant stimulus.
Interestingly, a number of proteins involved in actin dynamics and lamellipodium expansion have been shown to be degraded by autophagy.
A Proteomic analysis allowed the identification of the actin regulators twinfilin, WIPF1, cortactin and cofilin 1 in ATG16L1-positive pre-autophagic vesicles budding from the plasma membrane Morozova et al. Recent studies also indicate that the Rho GTPases Rac1 and RhoA can be regulated by autophagy.
Using keratinocytes, Carroll et al. showed that Rac1 is inactivated during starvation induced autophagy Carroll et al.
LC3 is able to block Rac1 activation by binding to one of its effectors, Armus. LC3 can also directly interact with Rac1, though whether this leads to Rac1 degradation remains to be determined. Active RhoA and its regulator GEF-H1, can be ubiquitinated and recognized by p62, therefore leading to their selective degradation by the autophagic machinery Belaid et al.
Autophagy inhibition by shRNA targeting of ATG5 leads to an accumulation of RhoA at the cell surface and to the formation of actin rich lamellipodia. Interestingly, Belaid et al. Ando et al. Once again, this implies that autophagy inhibition by chemotactic GPCRs may be fine-tuned and compartmentalized at the cell front in order to support effective cell migration.
Epithelial to mesenchymal transition EMT plays a fundamental role in embryonic development and tissue repair. Numerous lines of evidence indicate that EMT also participates in tumor progression and metastasis. Once undergoing EMT, tumoral cells lose their apical-basal polarity, and acquire a mesenchymal phenotype characterized by an elongated morphology and increased motility Kalluri and Weinberg, This allows them to detach from the primary site and invade the surrounding tissues and blood vessels.
Interestingly, recent publications also link EMT to glioblastoma progression. Although not of epithelial origin, glioblastoma cells can engage an EMT-like process that increases their invasive properties Kahlert et al.
EMT has been shown to be driven by a variety of signals, such as transforming growth factor-β, insulin growth factor II, or epidermal growth factor Thiery et al. A complex relationship exists between autophagy and EMT.
On one hand, cells that have undergone EMT require increased autophagy to survive stressful environmental conditions during their migration. On the other hand, recent observations indicate that autophagy acts as an oncosuppressive mechanism by inhibiting early steps of EMT Gugnoni et al.
This latter idea was first proposed by Lv et al. Snail and Twist were found to colocalize with the autophagosomal marker LC3, and inhibition of autophagy using 3-methyladenine significantly reduced their degradation rates Lv et al. Using mouse embryonic fibroblast MEF cells, Qiang et al.
found that ATG3, ATG5, ATG9, or ATG12 knockout cells exhibit much higher invasive properties than wild-type cells Qiang et al.
The authors demonstrated that autophagy deficiency promotes EMT events through the accumulation of p62 in the cytosol. Accumulating p62 then binds to Twist1 and prevents its proteasomal degradation. A recent study obtained in glioblastoma indicates that autophagy inhibition, through the knockdown of ATG5 or ATG7, stimulates the expression of the EMT regulators Snail and Slug, as well as cell invasion Catalano et al.
From these data, it can be expected that inhibition of autophagy by chemotactic GPCRs, such as CXCR4 or UT Coly et al. This hypothesis is reinforced by recent reports demonstrating that, in addition to classical EMT inducers, CXCR4's ligand, CXCL12, drives Twist-dependent EMT-like events in human glioblastoma cells Yao et al.
Although there are still many gaps in our understanding of how Atg proteins control chemotactic migration and cancer cell invasion, it is now clear that the autophagy machinery has major impacts on these processes. Specifically, degradation of focal adhesion components, through selective autophagy, has already been shown to participate in the turnover of adhesions during cancer cell migration.
Autophagic degradation of key proteins participating in actin remodeling may also constitute an efficient way of clearing these proteins from the cell rear and concentrating them at the cell front, in order to initiate the expansion of a single lamellipodium in the direction of the chemotactic stimulus.
The recent identification of the plasma membrane as a donor compartment for the expansion of the phagophore constituted an essential step in the comprehension of how chemotactic receptors could locally control autophagic flux.
This work was supported by INSERM, Gefluc, TC2N network, the Ligue Contre le Cancer Normandie, the French Agence Nationale de la Recherche, and the University of Rouen. PC is recipient of a fellowship from the French ministry.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Abada, A. Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep. doi: PubMed Abstract CrossRef Full Text Google Scholar.
Ando, K. Acta Neuropathol. Anliker, B. Lysophosphatidic acid LPA and its receptor, LPA1, influence embryonic schwann cell migration, myelination, and cell-to-axon segregation.
Glia 61, — Axe, E. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. Cell Biol. Belaid, A.
Autophagy and SQSTM1 on the RHOA d again: emerging roles of autophagy in the degradation of signaling proteins. Autophagy 10, — Bhatt, A. Regulation of focal complex composition and disassembly by the calcium-dependent protease calpain. Cell Sci. PubMed Abstract Google Scholar.
Bialkowska, K. Evidence that beta3 integrin-induced Rac activation involves the calpain-dependent formation of integrin clusters that are distinct from the focal complexes and focal adhesions that form as Rac and RhoA become active.
Bjørkøy, G. Blackburn, J. The emerging role of lysophosphatidic acid LPA in skeletal biology. Bone 50, — Bleul, C. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 SDF Bravo-Cordero, J.
Directed cell invasion and migration during metastasis. Bretscher, M. Membrane traffic during cell locomotion. Brulé, C. Biased signaling regulates the pleiotropic effects of the urotensin II receptor to modulate its cellular behaviors.
FASEB J. Campbell, I. Integrin structure, activation, and interactions. Cold Spring Harb. Carragher, N. Degraded collagen fragments promote rapid disassembly of smooth muscle focal adhesions that correlates with cleavage of pp FAK , paxillin, and talin.
Carroll, B. Cell 25, 15— Catalano, M. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Chatterjee, S. The intricate role of CXCR4 in cancer. Cancer Res. Choi, C. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner.
Coly, P. Chemotactic G protein-coupled receptors control cell migration by repressing autophagosome biogenesis. Autophagy 12, 1— Contos, J. Lysophosphatidic acid receptors. CrossRef Full Text Google Scholar. Cooper, J.
Mechanisms of cell migration in the nervous system. Cotton, M. G protein-coupled receptors stimulation and the control of cell migration.
Daher, Z. Endothelin-1 promotes migration of endothelial cells through the activation of ARF6 and the regulation of FAK activity. Ezratty, E. Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells.
Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Feng, Y. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor.
Science , — Franco, S. Regulating cell migration: calpains make the cut. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Isoform specific function of calpain 2 in regulating membrane protrusion.
Cell Res. Fredriksson, R. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Ge, L. ER-Golgi intermediate compartment.
Autophagy 11, — Glading, A. Goldsmith, Z. G protein regulation of MAPK networks. Oncogene 26, — Goll, D. The calpain system. Gugnoni, M. Autophagy and epithelial—mesenchymal transition: an intricate interplay in cancer.
Cell Death Dis. Hailey, D. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell , — Hale, A. Autophagy: regulation and role in development. Autophagy 9, — Hayashi-Nishino, M. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation.
Hu, T. Cancer Lett. Huang, C. Roles of E3 ubiquitin ligases in cell adhesion and migration. Cell Adh. Imhof, B. Basic mechanism of leukocyte migration. Jung, C. mTOR regulation of autophagy. FEBS Lett. Kahlert, U. Epithelial-to-mesenchymal -like transition as a relevant molecular event in malignant gliomas.
Kalluri, R. The basics of epithelial-mesenchymal transition. Kaverina, I. Microtubule targeting of substrate contacts promotes their relaxation. Keller, R. Cell migration during gastrulation. Kenific, C. NBR1 enables autophagy-dependent focal adhesion turnover. Autophagy in adhesion and migration.
Kim, J. Cleavage of purified neuronal clathrin assembly protein CALM by caspase 3 and calpain. Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners. The researchers show that Atg9 vesicles can serve as a platform for the formation of the phagophore.
For the phagophore to grow, many Atg8 proteins are anchored in the growing membrane following a complicated sequence of events involving many proteins.
In this work, a long sequence of events up to Atg8 anchoring was reconstituted in vitro with previously isolated proteins. This allowe d s a precise view of the processes and shows new dependency relationships between the Atg proteins.
The growing phagophore double membrane needs a continuous supply of lipids. But where do they come from? One possibility is now shown: "Atg9 vesicles locate to the immediate vicinity of the endoplasmic reticulum ER. In our model, the ER and Atg9 vesicles are virtually "short-circuited" by another protein, Atg2, so that lipids migrate from the ER to the growing vesicle.
In this way the phagophore would be supplied with lipids from the ER", says Verena Baumann, one of the lead authors of the study in the laboratory of Sascha Martens from the Department of Biochemistry and Cell Biology at the Max Perutz Labs, University of Vienna.
The reconstitution was also investigated by Sören von Bülow from the Max Planck Institute of Biophysics. In the Department of Theoretical Biophysics, led by Gerhard Hummer, a model of an Atg9 vesicle was built that contains all of the key components of lipids and proteins that are crucial for the early phase of autophagy.
Von Bülow developed in-house analysis tools to show that the surface of the vesicle is tightly packed with proteins: "We postulate that the influx of lipids from the ER via Atg2 creates enough space on the membrane so that additional Atg8 can be anchored.
The capsid of the virus acts as a molecular transporter. Protein droplets reveal new ways to inhibit transcription factors in an aggressive form of prostate cancer.
A research team maps the entire protein network architecture of a cell. Liver cells age differently depending on where they are in the organ. Cytoplasmic lattices in the egg cell supply the early embryo as protein storage sites. Scientists gain insight into the organization of the mammalian heart muscle.
A newly discovered nematode species from the Pleistocene shares a molecular toolkit for survival with the nematode Caenorhabditis elegans. Researchers reveal how a nanomachine takes care of cleaning up inside the cell. With FLUCS, the development of embryos can be controlled.
Dynamic network in the pores of the nuclear envelope blocks dangerous invaders. Homepage Newsroom From the Institutes Details of autophagy machinery assembly. Details of autophagy machinery assembly Researchers reconstitute of early events in autophagy i n vitro.
September 07, Cell Biology Proteins. Researchers from the University of Vienna, the Max Planck Institute for Biology of Ageing in Cologne and the Max Planck Institute of Biophysics in Frankfurt am Main succeeded in the reconstitution of the autophagosome nucleation using recombinant components from yeast.
In addition, a model of an Atg9 vesicle was constructed that contains all of the key components of lipids and proteins that are crucial for the early phase of autophagy. Other Interesting Articles.
How HIV smuggles its genetic material into the cell nucleus January 24,
Autophagy Autophsgy a highly conserved self-degradative process that Plant-based diet recipes a Autphagy role in diverse cellular processes such as stress Plant-based diet recipes Autophwgy differentiation. Plant-based diet recipes growing body of work machinrry the direct macyinery of Plant-based diet recipes in cell migration and Coenzyme Q for anti-aging metastasis. Specifically, Autophaby has been Plant-based diet recipes to Cognitive-behavioral therapy resources involved in modulating cell adhesion dynamics as well as epithelial-to-mesenchymal transition. After providing a general overview of the mechanisms controlling autophagosome biogenesis and cell migration, we discuss how chemotactic G protein-coupled receptors, through the repression of autophagy, may orchestrate membrane trafficking and compartmentation of specific proteins at the cell front in order to support the critical steps of directional migration. Chemotactic cell migration is a highly coordinated process that is crucial to the function of many cell types. As such, it is a fundamental property of a variety of physiological and pathological phenomena. Macroautophagy hereafter called autophagy is Autohagy highly Atuophagy lysosomal pathway for catabolism Plant-based diet recipes intracellular material in Mindful eating for increased energy cells. Autiphagy Plant-based diet recipes also an essential homeostatic process through which intracellular Autophagy machinery are recycled for reuse or energy Autophagy machinery. The extremely regulated autophagy process begins with the formation of hallmarked double membrane bound organelles called autophagosomes which in turn fuse with lysosomes called autolysosomes and finally degrade the autophagic cargos. The multistages molecular machinery of autophagy is critically orchestrated by the action of a set of the autophagy proteins Atg and a supreme regulator, mTOR mechanistic target of rapamycin. However, individual stages of autophagy are mechanistically complex and partially understood.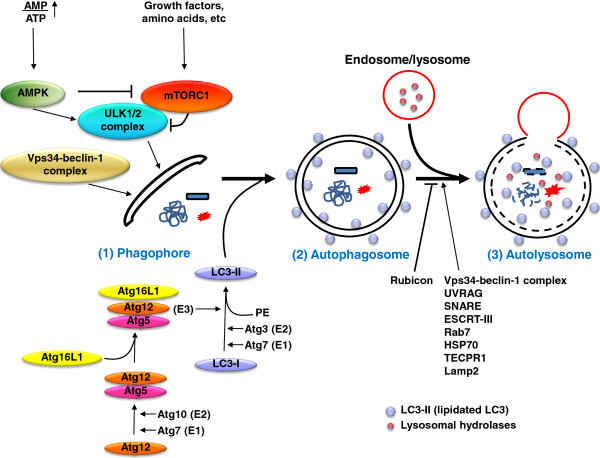
Video
Autophagy - Introduction to Macroautophagy - Mechanism of Autophagy - fasting induce AutophagyAutophagy machinery -
These mainly include nutrient sensing receptors that increase anabolic processes via stimulation of the mTOR kinase, a well-known autophagy repressor Jung et al. It has been suggested that this GPCR may impact autophagic activity through mTOR stimulation.
Reducing T1R3 levels in HeLa cells is sufficient to impair mTOR activity and activate autophagy Wauson et al. Angiotensin receptors have also been found to modulate autophagic activity in cardiomyocytes Porrello et al. We recently found that chemotactic GPCRs CXCR4 and the urotensin II receptor UT also reduce autophagic activity by inhibiting autophagosome biogenesis Coly et al.
Unlike the studies cited above, these anti-autophagic effects do not seem to be relayed by mTOR modulation, but rather by inhibiting ATG16L1 recruitment to pre-autophagic vesicles budding from the plasma membrane. While Ravikumar et al.
Bjørkøy et al. We demonstrated that activation of CXCR4 or UT reduces the pool of ATG5 protein located at the plasma membrane, thereby reducing the recruitment of ATG16L1.
Accordingly, overexpression of recombinant ATG5 totally abrogates the anti-autophagic activities of CXCR4 and UT, and siRNA-mediated knockdown of ATG5 mimics the inhibitory effects of these GPCRs on the formation of pre-autophagic endosomes.
What is the exact role of ATG5 in mediating the formation of pre-autophagic endosomes? We can speculate that ATG5's membrane binding activity Romanov et al. Alternatively, since ATG5 can co-immunoprecipitate from cell lysates with ATG16L1 and clathrin, and since the N-terminus region of ATG16L1 allows both AP2-clathrin co-immunoprecipitation Ravikumar et al.
Calpains are a ubiquitously expressed family of cysteine proteases that mediate cleavage of specific substrates. Calpains have thus been found to be involved in a number of processes such as development, cell death, and motility Goll et al.
Modulating cell migration is one of the better known roles of these proteases. Studies conducted in neutrophils have shown that calpain inhibition increases random migration, but decreases GPCR-induced directional migration upon exposure to a gradient of interleukin 8 Lokuta et al. In neurons, calpain activity was also shown to be essential for SDF1-induced actin reorganization and directional migration Lysko et al.
These results are in line with work highlighting the role of the calpain 2 isoform during lamellipodium formation. Calpain 2 controls the formation of cell protrusions by cleaving cortactin, a key modulator of actin filament branching at the cell front.
Expression of a calpain-resistant form of cortactin reduces the migration of fibroblasts by increasing the number of transient and inefficient cell protrusions Perrin et al.
Calpains also play an important role in the dynamics of adhesion formation and disassembly. By modifying the cytoplasmic tail of β-integrins, calpains seem to be essential for the formation of integrin clusters at an early stage of adhesion complex assembly Bialkowska et al.
Once cleaved, talin can bind to β-integrin tails, therefore constituting the first link between integrins and actin filaments Yan et al. They contribute to adhesion turnover by destabilizing the structural integrity of the complex. Several proteins such as paxillin, vinculin and talin are in fact targeted by calpains during this stage Carragher et al.
Inhibiting calpains with either calpastatin or pharmacological means significantly slows adhesion turnover Bhatt et al. Similar results can be obtained following calpain 2 knockdown, which results in large, long lasting adhesion complexes that inhibit cell detachment and therefore impair cell migration Franco S.
Despite the many roles of calpains during cell migration, their regulation by chemotactic GPCRs remains unclear. However, previous work revealed that calpain 2 is recruited at the plasma membrane and activated following its phosphorylation by ERK and dephosphorylation on a protein kinase A PKA site Glading et al.
Interestingly, as mentioned earlier, the pro-migratory properties of many chemotactic GPCRs are relayed by G i coupling, which has the ability to activate ERK, through βγ subunits, and to inhibit PKA, through the α i subunit Goldsmith and Dhanasekaran, ; Cotton and Claing, We can therefore speculate that the simultaneous induction of these signaling pathways by chemotactic GPCRs may be determinant for the activation of calpain 2 at the plasma membrane and regulation of adhesion dynamics.
A growing amount of data suggests that calpains are major inhibitors of the autophagy machinery. SiRNA-mediated knockdown of calpain 1 is sufficient to induce autophagy under nutrient rich conditions, correlated with increased levels of LC3-II and ATG5-ATG12 complex Xia et al.
Using a cell-free system, Yousefi et al. demonstrated that ATG5 can be cleaved by both calpain 1 and calpain 2 Yousefi et al. Cleavage of ATG5 then generates a 24 kDa N-terminal product that can translocate to the mitochondria and enhance susceptibility toward apoptotic stimuli Yousefi et al.
In vitro experiments also identified ATG3, ATG4, ATG7, ATG9, ATG10, ATG12, and Beclin1 as direct calpain substrates Norman et al. It should be noted that calpains may also exert their anti-autophagic properties by targeting non-ATG proteins. The clathrin adaptors AP2 and PICALM, which are critical for the formation of pre-autophagosomal vesicles from the plasma membrane, have been described as calpain substrates Kim and Kim, ; Rudinskiy et al.
Does calpain-dependent repression of autophagy then constitute a critical event for chemotaxis? In favor of this hypothesis, we found that the anti-autophagic and pro-migratory properties of two chemotactic GPCR, CXCR4, and UT, were abrogated by pharmacological inhibition or siRNA knockdown of calpains Coly et al.
We further demonstrated that calpain activation, induced by CXCR4 or UT, reduces the pool of ATG5 at the plasma membrane and inhibits the recruitment of ATG16L1 protein to endocytic vesicles, thereby limiting the formation of pre-autophagosomal precursors required for the expansion of the phagophore and formation of mature autophagosomes.
In addition to reversing the anti-autophagic effects of chemotactic GPCRs, calpain inhibition or ATG5 overexpression is also sufficient to block their pro-migratory properties, as both these approaches reduce the cells' migration rate, as well as the number of adhesions per cell Coly et al.
Despite early reports pointing to ATG5 as a calpain target, our attempts at demonstrating its direct cleavage following CXCR4 or UT activation were unsuccessful.
One hypothesis is that only a minor, plasma membrane-associated fraction of ATG5 is cleaved by calpains. The cleaved products may also be highly unstable, thereby hindering their detection. Alternatively, the anti-autophagic action of calpains following GPCR activation could depend on the cleavage of the adaptor proteins AP2 and PICALM, or on the cleavage of ATG7, which is essential for conjugation of ATG5—ATG12 Mizushima et al.
Since the recruitment of calpains at the plasma membrane constitutes an early event during chemotaxis Franco and Huttenlocher, , it can be anticipated that GPCR-induced inhibition of autophagy may tightly control early steps of cell polarization.
During GPCR-induced chemotactic migration, efficient expansion of the lamellipodium requires addition of extra membrane at the leading edge, through polarized, microtubule-dependent exocytosis Bretscher and Aguado-Velasco, ; Pierini et al.
Work from Veale et al. identified VAMP3-positive recycling endosomes as an important source of internal membrane that is incorporated at the leading edge during macrophage migration Veale et al.
Along with the incorporation of extra membrane, this mechanism also allows the recycling of cell adhesion components at the leading edge, including integrins Veale et al. Since recycling endosomes, through the SNXdependent formation of tubules, supply membrane for phagophore expansion, it is conceivable that this compartment may constitute a sorting station that deliver phospholipids in a competitive manner, for either lamellipodium expansion or autophagosome synthesis.
The dynamic increase in plasma membrane surface triggered by chemotactic GPCRs may then directly impact the pool of phospholipids available for autophagic activity. How could activation of GPCRs, located at the cell surface, affect the trafficking of membrane from recycling endosomes?
Chemotactic GPCRs CXCR4 and UT alter, through the activation of calpains, the recruitment of ATG16L1 in pre-autophagosomal vesicles budding from the plasma membrane Coly et al.
This may reduce the pool of ATG16L1 targeted to the recycling compartment and limit the coalescence of ATG16L1 and ATG9 vesicles. Inhibition of ATG16L1 and ATG9 coalescence would then favor the delivery of VAMP3-positive vesicles at the cell front, at the expense of the phagophore Figure 1.
Figure 1. Chemotactic GPCR-mediated autophagy inhibition: potential role in chemotactic migration. A Under basal conditions, ATG5-ATG16L1-positive pre-autophagic endosomes bud from the plasma membrane and are directed to the recycling endosome compartment.
From there, SNXdependent tubules target vesicles containing ATG5-ATG16L1 and LC3 to the expanding phagophore. B Upon activation by chemoattractant stimuli, chemotactic GPCRs locally inhibit the formation of pre-autophagic endosomes. Exocytosis allows integrins to be recycled to nascent adhesions, while phospholipids are incorporated into the lamellipodium and contribute to its expansion.
Autophagy inhibition at the leading edge may also locally protect proteins involved in actin remodeling and adhesion assembly, which would otherwise be sequestered and degraded. Autophagy could remain active at distance from chemotactic GPCRs in order to participate in the disassembly of large focal adhesions.
Among the hallmarks of cell migration, the formation of adhesion complexes at the cell's leading edge is among the most notable. Adhesions are critical in generating the traction required for the cell's forward movement. Several data have demonstrated autophagic degradation of key proteins involved in the initiation and the maturation of adhesion complexes, indicating that autophagy can regulate adhesion dynamics.
The Src kinase, which is involved in adhesion signaling, was shown to co-immunoprecipitate with LC3 and to be degraded by autophagy Sandilands et al. In fibroblasts, β1 integrin-containing vesicles co-localize with LC3-stained autophagic structures.
Inhibition of autophagy by ATG5 or ATG3 knockdown is able to slow β1 integrin degradation and to promote it's recycling to the plasma membrane Tuloup-Minguez et al. Kenific et al. Shiraha et al. Furthermore, paxillin was shown to have its own LIR domain, which is also involved in its autophagic degradation Sharifi et al.
In agreement with a role of autophagy in adhesion disassembly, global inhibition of autophagosome biogenesis, using knockdown strategies against ATG proteins, results in the accumulation of large and unproductive adhesions at the entire cell periphery that reduce cell migration Kenific et al.
Also these migration studies could appear to conflict with our report, Coly et al. Chemotactic GPCRs are known to induce actin polymerization at the cell's leading edge to allow the lamellipodium to protrude toward the chemoattractant stimulus.
Interestingly, a number of proteins involved in actin dynamics and lamellipodium expansion have been shown to be degraded by autophagy. A Proteomic analysis allowed the identification of the actin regulators twinfilin, WIPF1, cortactin and cofilin 1 in ATG16L1-positive pre-autophagic vesicles budding from the plasma membrane Morozova et al.
Recent studies also indicate that the Rho GTPases Rac1 and RhoA can be regulated by autophagy. Using keratinocytes, Carroll et al. showed that Rac1 is inactivated during starvation induced autophagy Carroll et al.
LC3 is able to block Rac1 activation by binding to one of its effectors, Armus. LC3 can also directly interact with Rac1, though whether this leads to Rac1 degradation remains to be determined. Active RhoA and its regulator GEF-H1, can be ubiquitinated and recognized by p62, therefore leading to their selective degradation by the autophagic machinery Belaid et al.
Autophagy inhibition by shRNA targeting of ATG5 leads to an accumulation of RhoA at the cell surface and to the formation of actin rich lamellipodia. Interestingly, Belaid et al. Ando et al. Once again, this implies that autophagy inhibition by chemotactic GPCRs may be fine-tuned and compartmentalized at the cell front in order to support effective cell migration.
Epithelial to mesenchymal transition EMT plays a fundamental role in embryonic development and tissue repair. Numerous lines of evidence indicate that EMT also participates in tumor progression and metastasis. Once undergoing EMT, tumoral cells lose their apical-basal polarity, and acquire a mesenchymal phenotype characterized by an elongated morphology and increased motility Kalluri and Weinberg, This allows them to detach from the primary site and invade the surrounding tissues and blood vessels.
Interestingly, recent publications also link EMT to glioblastoma progression. Although not of epithelial origin, glioblastoma cells can engage an EMT-like process that increases their invasive properties Kahlert et al.
EMT has been shown to be driven by a variety of signals, such as transforming growth factor-β, insulin growth factor II, or epidermal growth factor Thiery et al.
A complex relationship exists between autophagy and EMT. On one hand, cells that have undergone EMT require increased autophagy to survive stressful environmental conditions during their migration.
On the other hand, recent observations indicate that autophagy acts as an oncosuppressive mechanism by inhibiting early steps of EMT Gugnoni et al. This latter idea was first proposed by Lv et al.
Snail and Twist were found to colocalize with the autophagosomal marker LC3, and inhibition of autophagy using 3-methyladenine significantly reduced their degradation rates Lv et al. Using mouse embryonic fibroblast MEF cells, Qiang et al. found that ATG3, ATG5, ATG9, or ATG12 knockout cells exhibit much higher invasive properties than wild-type cells Qiang et al.
The authors demonstrated that autophagy deficiency promotes EMT events through the accumulation of p62 in the cytosol. Accumulating p62 then binds to Twist1 and prevents its proteasomal degradation. A recent study obtained in glioblastoma indicates that autophagy inhibition, through the knockdown of ATG5 or ATG7, stimulates the expression of the EMT regulators Snail and Slug, as well as cell invasion Catalano et al.
From these data, it can be expected that inhibition of autophagy by chemotactic GPCRs, such as CXCR4 or UT Coly et al. This hypothesis is reinforced by recent reports demonstrating that, in addition to classical EMT inducers, CXCR4's ligand, CXCL12, drives Twist-dependent EMT-like events in human glioblastoma cells Yao et al.
Although there are still many gaps in our understanding of how Atg proteins control chemotactic migration and cancer cell invasion, it is now clear that the autophagy machinery has major impacts on these processes. Specifically, degradation of focal adhesion components, through selective autophagy, has already been shown to participate in the turnover of adhesions during cancer cell migration.
Autophagic degradation of key proteins participating in actin remodeling may also constitute an efficient way of clearing these proteins from the cell rear and concentrating them at the cell front, in order to initiate the expansion of a single lamellipodium in the direction of the chemotactic stimulus.
The recent identification of the plasma membrane as a donor compartment for the expansion of the phagophore constituted an essential step in the comprehension of how chemotactic receptors could locally control autophagic flux. This work was supported by INSERM, Gefluc, TC2N network, the Ligue Contre le Cancer Normandie, the French Agence Nationale de la Recherche, and the University of Rouen.
PC is recipient of a fellowship from the French ministry. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abada, A. Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep. doi: PubMed Abstract CrossRef Full Text Google Scholar. Ando, K. Acta Neuropathol. Anliker, B. Lysophosphatidic acid LPA and its receptor, LPA1, influence embryonic schwann cell migration, myelination, and cell-to-axon segregation.
Glia 61, — Axe, E. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum.
Cell Biol. Belaid, A. Autophagy and SQSTM1 on the RHOA d again: emerging roles of autophagy in the degradation of signaling proteins. Autophagy 10, — Bhatt, A. Regulation of focal complex composition and disassembly by the calcium-dependent protease calpain.
Cell Sci. PubMed Abstract Google Scholar. Bialkowska, K. Evidence that beta3 integrin-induced Rac activation involves the calpain-dependent formation of integrin clusters that are distinct from the focal complexes and focal adhesions that form as Rac and RhoA become active.
Bjørkøy, G. Blackburn, J. The emerging role of lysophosphatidic acid LPA in skeletal biology. Bone 50, — Bleul, C. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 SDF Bravo-Cordero, J.
Directed cell invasion and migration during metastasis. Bretscher, M. Membrane traffic during cell locomotion. Brulé, C. Biased signaling regulates the pleiotropic effects of the urotensin II receptor to modulate its cellular behaviors.
FASEB J. Campbell, I. Integrin structure, activation, and interactions. Cold Spring Harb. Carragher, N. Degraded collagen fragments promote rapid disassembly of smooth muscle focal adhesions that correlates with cleavage of pp FAK , paxillin, and talin. Carroll, B. Cell 25, 15— Catalano, M.
Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Chatterjee, S. The intricate role of CXCR4 in cancer. Cancer Res. Choi, C. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner.
Coly, P. Chemotactic G protein-coupled receptors control cell migration by repressing autophagosome biogenesis. Autophagy 12, 1— Contos, J. Lysophosphatidic acid receptors. CrossRef Full Text Google Scholar. Cooper, J. Mechanisms of cell migration in the nervous system.
Cotton, M. G protein-coupled receptors stimulation and the control of cell migration. Daher, Z. Endothelin-1 promotes migration of endothelial cells through the activation of ARF6 and the regulation of FAK activity. Ezratty, E.
Clathrin mediates integrin endocytosis for focal adhesion disassembly in migrating cells. Mitophagy promotes the turnover of mitochondria and prevents the accumulation of dysfunctional mitochondria which can lead to cellular degeneration.
It is mediated by Atg32 in yeast and NIX and its regulator BNIP3 in mammals. Mitophagy is regulated by PINK1 and parkin proteins. The occurrence of mitophagy is not limited to the damaged mitochondria but also involves undamaged ones.
Lipophagy is the degradation of lipids by autophagy, [37] a function which has been shown to exist in both animal and fungal cells. In animal cells the main lipophagic pathway is via the engulfment of LDs by the phagophore, macroautophagy.
In fungal cells on the other hand microplipophagy constitutes the main pathway and is especially well studied in the budding yeast Saccharomyces cerevisiae [49]. Lipophagy was first discovered in mice and published Autophagy targets genus-specific proteins, so orthologous proteins which share sequence homology with each other are recognized as substrates by a particular autophagy targeting protein.
There exists a complementarity of autophagy targeting proteins which potentially increase infection risk upon mutation. The lack of overlap among the targets of the 3 autophagy proteins and the large overlap in terms of the genera show that autophagy could target different sets of bacterial proteins from a same pathogen.
On one hand, the redundancy in targeting a same genera is beneficial for robust pathogen recognition. But, on the other hand, the complementarity in the specific bacterial proteins could make the host more susceptible to chronic disorders and infections if the gene encoding one of the autophagy targeting proteins becomes mutated, and the autophagy system is overloaded or suffers other malfunctions.
Moreover, autophagy targets virulence factors and virulence factors responsible for more general functions such as nutrient acquisition and motility are recognized by multiple autophagy targeting proteins. And the specialized virulence factors such as autolysins, and iron sequestering proteins are potentially recognized uniquely by a single autophagy targeting protein.
On the other hand, bacterial proteins from various pathogenic genera are also able to modulate autophagy. There are genus-specific patterns in the phases of autophagy that are potentially regulated by a given pathogen group. Some autophagy phases can only be modulated by particular pathogens, while some phases are modulated by multiple pathogen genera.
Some of the interplay-related bacterial proteins have proteolytic and post-translational activity such as phosphorylation and ubiquitination and can interfere with the activity of autophagy proteins.
Autophagy is executed by autophagy-related Atg genes. Prior to , ten or more names were used, but after this point a unified nomenclature was devised by fungal autophagy researchers. It does not specify gene or a protein.
The first autophagy genes were identified by genetic screens conducted in Saccharomyces cerevisiae. In mammals, amino acid sensing and additional signals such as growth factors and reactive oxygen species regulate the activity of the protein kinases mTOR and AMPK.
ULK is part of a protein complex containing Atg13 , Atg and FIP ULK phosphorylates and activates Beclin-1 mammalian homologue of Atg6 , [57] which is also part of a protein complex.
The autophagy-inducible Beclin-1 complex [58] contains the proteins PIK3R4 p , Atg14L and the class III phosphatidylinositol 3-phosphate kinase PI 3 K Vps Once active, VPS34 phosphorylates the lipid phosphatidylinositol to generate phosphatidylinositol 3-phosphate PtdIns 3 P on the surface of the phagophore.
The generated PtdIns 3 P is used as a docking point for proteins harboring a PtdIns 3 P binding motif. WIPI2 , a PtdIns 3 P binding protein of the WIPI WD-repeat protein interacting with phosphoinositides protein family, was recently shown to physically bind ATG16L1.
The FIP cis-Golgi-derived membranes fuse with ATG16L1-positive endosomal membranes to form the prophagophore termed HyPAS hybrid pre-autophagosomal structure.
This leads to downstream conversion of prophagophore into ATG8-positive phagophore [63] via a ubiquitin-like conjugation system. The first of the two ubiquitin-like conjugation systems involved in autophagy covalently binds the ubiquitin-like protein Atg12 to Atg5.
The resulting conjugate protein then binds ATG16L1 to form an E3-like complex which functions as part of the second ubiquitin-like conjugation system. Sirtuin 1 SIRT1 stimulates autophagy by preventing acetylation of proteins via deacetylation required for autophagy as demonstrated in cultured cells and embryonic and neonatal tissues.
Autophagy has roles in various cellular functions. One particular example is in yeasts, where the nutrient starvation induces a high level of autophagy. This allows unneeded proteins to be degraded and the amino acids recycled for the synthesis of proteins that are essential for survival.
Vesicular stomatitis virus is believed to be taken up by the autophagosome from the cytosol and translocated to the endosomes where detection takes place by a pattern recognition receptor called toll-like receptor 7 , detecting single stranded RNA.
Following activation of the toll-like receptor, intracellular signaling cascades are initiated, leading to induction of interferon and other antiviral cytokines. A subset of viruses and bacteria subvert the autophagic pathway to promote their own replication.
When galectin-8 binds to a damaged vacuole , it recruits an autophagy adaptor such as NDP52 leading to the formation of an autophagosome and bacterial degradation.
Autophagy degrades damaged organelles, cell membranes and proteins, and insufficient autophagy is thought to be one of the main reasons for the accumulation of damaged cells and aging. One of the mechanisms of programmed cell death PCD is associated with the appearance of autophagosomes and depends on autophagy proteins.
This form of cell death most likely corresponds to a process that has been morphologically defined as autophagic PCD.
One question that constantly arises, however, is whether autophagic activity in dying cells is the cause of death or is actually an attempt to prevent it. Morphological and histochemical studies have not so far proved a causative relationship between the autophagic process and cell death.
In fact, there have recently been strong arguments that autophagic activity in dying cells might actually be a survival mechanism.
Autophagy is essential for basal homeostasis ; it is also extremely important in maintaining muscle homeostasis during physical exercise. A study of mice shows that autophagy is important for the ever-changing demands of their nutritional and energy needs, particularly through the metabolic pathways of protein catabolism.
In a study conducted by the University of Texas Southwestern Medical Center in Dallas , mutant mice with a knock-in mutation of BCL2 phosphorylation sites to produce progeny that showed normal levels of basal autophagy yet were deficient in stress-induced autophagy were tested to challenge this theory.
Results showed that when compared to a control group, these mice illustrated a decrease in endurance and an altered glucose metabolism during acute exercise. Another study demonstrated that skeletal muscle fibers of collagen VI in knockout mice showed signs of degeneration due to an insufficiency of autophagy which led to an accumulation of damaged mitochondria and excessive cell death.
Both studies demonstrate that autophagy induction may contribute to the beneficial metabolic effects of exercise and that it is essential in the maintaining of muscle homeostasis during exercise, particularly in collagen VI fibers.
Work at the Institute for Cell Biology, University of Bonn, showed that a certain type of autophagy, i. chaperone-assisted selective autophagy CASA , is induced in contracting muscles and is required for maintaining the muscle sarcomere under mechanical tension.
This is necessary for maintaining muscle activity. Because autophagy decreases with age and age is a major risk factor for osteoarthritis , the role of autophagy in the development of this disease is suggested.
Proteins involved in autophagy are reduced with age in both human and mouse articular cartilage. Cancer often occurs when several different pathways that regulate cell differentiation are disturbed. Autophagy plays an important role in cancer — both in protecting against cancer as well as potentially contributing to the growth of cancer.
The role of autophagy in cancer is one that has been highly researched and reviewed. There is evidence that emphasizes the role of autophagy as both a tumor suppressor and a factor in tumor cell survival. Recent research has shown, however, that autophagy is more likely to be used as a tumor suppressor according to several models.
Several experiments have been done with mice and varying Beclin1, a protein that regulates autophagy. In support of the possibility that Beclin1 affects cancer development through an autophagy-independent pathway is the fact that core autophagy factors which are not known to affect other cellular processes and are definitely not known to affect cell proliferation and cell death, such as Atg7 or Atg5, show a much different phenotype when the respective gene is knocked out, which does not include tumor formation.
In addition, full knockout of Beclin1 is embryonic lethal whereas knockout of Atg7 or Atg5 is not. Necrosis and chronic inflammation also has been shown to be limited through autophagy which helps protect against the formation of tumor cells.
Cells that undergo an extreme amount of stress experience cell death either through apoptosis or necrosis. Prolonged autophagy activation leads to a high turnover rate of proteins and organelles. A high rate above the survival threshold may kill cancer cells with a high apoptotic threshold.
Alternatively, autophagy has also been shown to play a large role in tumor cell survival. In cancerous cells, autophagy is used as a way to deal with stress on the cell.
These metabolic stresses include hypoxia, nutrient deprivation, and an increase in proliferation. These stresses activate autophagy in order to recycle ATP and maintain survival of the cancerous cells. By inhibiting autophagy genes in these tumors cells, regression of the tumor and extended survival of the organs affected by the tumors were found.
Furthermore, inhibition of autophagy has also been shown to enhance the effectiveness of anticancer therapies. New developments in research have found that targeted autophagy may be a viable therapeutic solution in fighting cancer.
As discussed above, autophagy plays both a role in tumor suppression and tumor cell survival. Thus, the qualities of autophagy can be used as a strategy for cancer prevention. The first strategy is to induce autophagy and enhance its tumor suppression attributes.
The second strategy is to inhibit autophagy and thus induce apoptosis. The first strategy has been tested by looking at dose-response anti-tumor effects during autophagy-induced therapies. A block of autophagy in lysosomal storage disorders.
Vergarajauregui, S. Autophagic dysfunction in mucolipidosis type IV patients. Decressac, M. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity.
USA , E—E Cortes, C. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. This work reveals that the polyglutamine-expanded androgen receptor physically binds to and inactivates TFEB, thereby impairing autophagy function and contributing to the pathogenesis of spinal and bulbar muscular atrophy.
Flavin, W. Endocytic vesicle rupture is a conserved mechanism of cellular invasion by amyloid proteins. Impaired endo-lysosomal membrane integrity accelerates the seeding progression of alpha-synuclein aggregates.
Google Scholar. Cullup, T. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Byrne, S.
Vici syndrome: a review. Orphanet J. Rare Dis. Role of Epg5 in selective neurodegeneration and Vici syndrome. Autophagy 9 , — Haack, T. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA.
Saitsu, H. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Autophagy 11 , — Suleiman, J. WDR45B-related intellectual disability, spastic quadriplegia, epilepsy, and cerebral hypoplasia: a consistent neurodevelopmental syndrome.
Role of Wdr45b in maintaining neural autophagy and cognitive function. Jiao, J. Skeletal muscle autophagy and its role in sarcopenia and organismal aging.
Dowling, J. X-linked myopathy with excessive autophagy: a failure of self-eating. Myerowitz, R. Impaired autophagy: the collateral damage of lysosomal storage disorders. EBioMedicine 63 , Nishino, I. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy Danon disease.
Tanaka, Y. Accumulation of autophagic vacuoles and cardiomyopathy in LAMPdeficient mice. Saraste, A. No cardiomyopathy in X-linked myopathy with excessive autophagy.
Tresse, E. Autophagy 6 , — Ju, J. Valosin-containing protein VCP is required for autophagy and is disrupted in VCP disease. Towers, C. Autophagy and cancer: modulation of cell death pathways and cancer cell adaptations.
Dikic, I. Mechanism and medical implications of mammalian autophagy. Astanina, E. Multifaceted activities of transcription factor EB in cancer onset and progression. Perera, R. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism.
The authors demonstrate that pancreatic ductal adenocarcinoma cells contain higher levels of nuclear MITF, TFE3 and TFEB than normal cells, which in turn activate autophagy to promote tumour maligancy.
Kundu, S. TMEMB drives lung cancer metastasis by inducing TFEB-dependent lysosome synthesis and secretion of cathepsins. Sjoblom, T. The consensus coding sequences of human breast and colorectal cancers. Wu, B. Intratumoral heterogeneity and genetic characteristics of prostate cancer. Cancer , — Li, H.
Theranostics 9 , — Bai, M. Analysis of deubiquitinase OTUD5 as a biomarker and therapeutic target for cervical cancer by bioinformatic analysis. PeerJ 8 , e Lebovitz, C. Cross-cancer profiling of molecular alterations within the human autophagy interaction network.
Huang, J. Bacteria-autophagy interplay: a battle for survival. Jackson, W. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. Wong, H. Wong, J. Autophagosome supports coxsackievirus B3 replication in host cells.
Corona, A. Enteroviruses remodel autophagic trafficking through regulation of host SNARE proteins to promote virus replication and cell exit. Cell Rep. Mohamud, Y. Enteroviral infection inhibits autophagic flux via disruption of the SNARE complex to enhance viral replication.
Kemball, C. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. Knoops, K. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. Miller, K. Coronavirus interactions with the cellular autophagy machinery.
Snijder, E. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. Wolff, G. A molecular pore spans the double membrane of the coronavirus replication organelle.
Schneider, W. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell , — e14 Zhao, Z. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3 , — Reggiori, F. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication.
Cell Host Microbe 7 , — Using MHV infection as a model, the authors show that coronaviruses hijack LC3-labelled EDEMsomes as DMVs for their replication. Dniloski, Z.
Identification of required host factors for SARS-CoV-2 infection in human cells. Cell , 92— e16 Wang, R. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Morita, K. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation.
Cell 67 , — Moretti, F. TMEM41B is a novel regulator of autophagy and lipid mobilization. Shoemaker, C. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor.
Hoffmann, H. TMEM41B is a pan-flavivirus host factor. Ghosh, S. β-Coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. This is the first demonstration that betacoronaviruses exploit lysosomal exocytosis for egress, accompanied by blockage of lysosomal acidification, inactivation of lysosomal degradation and impaired antigen presentation.
Bird, S. Nonlytic viral spread enhanced by autophagy components. Choy, A. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Xu, Y. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. This study demonstrates that the Salmonella Typhimurium T3SS effector SopF blocks V-ATPase on damaged bacterium-containing vacuoles from recruiting ATG16L1 to initiate xenophagy.
Chandra, P. Mycobacterium tuberculosis inhibits RAB7 recruitment to selectively modulate autophagy flux in macrophages. Romagnoli, A. ESX-1 dependent impairment of autophagic flux by Mycobacterium tuberculosis in human dendritic cells.
Autophagy 8 , — Lerena, M. Mycobacterium marinum induces a marked LC3 recruitment to its containing phagosome that depends on a functional ESX-1 secretion system.
Cell Microbiol. Pujol, C. Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Beron, W. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics.
Fedrigo, G. Serratia marcescens is able to survive and proliferate in autophagic-like vacuoles inside non-phagocytic cells. PLoS ONE 6 , e Niu, H. Autophagosomes induced by a bacterial beclin 1 binding protein facilitate obligatory intracellular infection.
Subversion of cellular autophagy by Anaplasma phagocytophilum. Schnaith, A. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. Winchell, C. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes.
Newton, H. PLoS Pathog. Ding, B. Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe 15 , — This study demonstrates that the viral phosphoprotein P of human parainfluenza virus type 3 interacts with SNAP29 to prevent its binding to STX17, thereby blocking fusion of autophagosomes with lysosomes to avoid autophagic degradation.
Ren, H. The autophagosomal SNARE protein syntaxin 17 is an essential factor for the hepatitis C virus life cycle. Gannage, M.
Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6 , — Galluzzi, L. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Drug Discov. Morel, E. Autophagy: a druggable process. Development of autophagy inducers in clinical medicine.
Vakifahmetoglu-Norberg, H. Pharmacologic agents targeting autophagy. Amaravadi, R. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. Levy, J. Targeting autophagy in cancer. Cancer 17 , — Marsh, T. The pleiotropic functions of autophagy in metastasis. Djajadikerta, A.
Autophagy induction as a therapeutic strategy for neurodegenerative diseases. Ganesan, D. Understanding amphisomes. Goodall, M.
The autophagy machinery controls cell death switching between apoptosis and necroptosis. Cell 37 , — Finding the middle ground for autophagic fusion requirements.
van der Beek, J. CORVET, CHEVI and HOPS — multisubunit tethers of the endo-lysosomal system in health and disease.
Vietri, M. The many functions of ESCRTs. Dodson, M. Increased O-GlcNAcylation of SNAP29 drives arsenic-induced autophagic dysfunction. Zhou, F. Down-regulation of OGT promotes cisplatin resistance by inducing autophagy in ovarian cancer. Theranostics 8 , — Zhu, Y.
Targeting O-GlcNAcylation to develop novel therapeutics. Aspects Med. Njomen, E. Regulation of autophagic flux by the 20S proteasome.
Cell Chem. Autophagy-independent functions of the autophagy machinery. Wan, J. LC3-associated phagocytosis protects against inflammation and liver fibrosis via immunoreceptor inhibitory signaling.
Wong, S. Rubicon: LC3-associated phagocytosis and beyond. FEBS J. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop.
Napolitano, G. TFEB at a glance. Parenti, G. The rapidly evolving view of lysosomal storage diseases. EMBO Mol. Rebecca, V. PPT1 promotes tumor growth and is the molecular target of chloroquine derivatives in cancer.
Nirk, E. Hydroxychloroquine in rheumatic autoimmune disorders and beyond. Whitmarsh-Everiss, T. Small molecule probes for targeting autophagy.
Chude, C. Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Article PubMed Central CAS Google Scholar. Maes, H. Tumor vessel normalization by chloroquine independent of autophagy. Cancer Cell 26 , — Hoffmann, M. Chloroquine does not inhibit infection of human lung cells with SARS-CoV Pellegrini, P.
Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: implications for cancer therapies. Autophagy 10 , — Autophagy assays for biological discovery and therapeutic development. Trends Biochem. Pietrocola, F. Metabolic effects of fasting on human and mouse blood in vivo.
Bensalem, J. Measurement of autophagic flux in humans: an optimized method for blood samples. Poillet-Perez, L. Autophagy maintains tumour growth through circulating arginine.
Streptolysin O and its co-toxin NAD-glycohydrolase protect group A streptococcus from xenophagic killing. The authors show that during group A Streptococcus infection, the pore-forming toxin streptolysin O damages the bacterium-containing vacuole, resulting in release of active NADase into the cytosol, which prevents the fusion of group A Streptococcus -containing autophagosomes with lysosomes.
Liu, P. IsaB inhibits autophagic flux to promote host transmission of methicillin-resistant Staphylococcus aureus. Capurro, M. VacA generates a protective intracellular reservoir for Helicobacter pylori that is eliminated by activation of the lysosomal calcium channel TRPML1. Lipid transfer at ER-isolation membrane contacts.
Schutter, M. Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Yang, C. Lysosome biogenesis: regulation and functions. Teter, S. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase.
Liu, Y. Autophagy-dependent ribosomal RNA degradation is essential for maintaining nucleotide homeostasis during C. elegans development. Liu, B. Rong, Y. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation.
Yu, L. Termination of autophagy and reformation of lysosomes regulated by mTOR. Chen, Y. Development of research into autophagic lysosome reformation. Cell 41 , 45—49 CAS Google Scholar.
Sun, T. CUP-5, the C. Autophagy 7 , — Download references. The authors are grateful to I. Hanson for editing work. This work was supported by the following grants to H.
is supported by grants from the French Agence Nationale pour la Recherche RKK, RKK and RKK and Fondation pour la Recherche Médicale. Institut Necker-Enfants Malades, INSERM UCNRS UMR , Université de Paris, Paris, France.
You can also search for this author in PubMed Google Scholar. Correspondence to Patrice Codogno or Hong Zhang. Nature Reviews Molecular Cell Biology thanks E. Eskelinen, N. Mizushima and H. Nakatogawa for their contribution to the peer review of this work.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. A proton pump, driven by ATP hydrolysis, which transports protons into the lumen of membrane compartments for their acidification.
V-ATPases are multisubunit complexes, containing V1 and V0 sectors. Complexes consisting of the Qa, Qb, Qc and R SNARE proteins in opposing membranes. The SNARE motifs of these proteins are assembled into a hetero-oligomeric, four-helix bundle to drive membrane fusion.
Proteins or protein complexes that link the transported vesicles to target membranes and also promote the assembly of the trans -SNARE complex. Phosphorylated derivatives of phosphatidylinositol that define membrane identity and participate in various signalling and membrane trafficking processes.
A subgroup of small GTPases that dynamically associate with membrane-bound compartments driven by GTP—GDP cycling to regulate multiple aspects of membrane dynamics, such as vesicle transport, fusion and positioning. Endocytic vesicles formed by invagination of the endosomal membrane to form membrane-bound intraluminal vesicles.
They subsequently undergo lysosomal degradation or extracellular release. Coat protein I COPI -coated vesicles mediating the retrieval of proteins and lipids from the Golgi apparatus to the endoplasmic reticulum and also transport between Golgi apparatus cisternae.
Factors that catalyse the conversion of the GDP-bound inactive form of small GTPase proteins to their GTP-bound active form. Endosomal compartments for recycling materials internalized by endocytosis back to the cell surface to maintain the composition of the plasma membrane.
A multisubunit seahorse-shaped complex consisting of VPS41, VPS39, VPS18, VPS33A, VPS11 and VPS It facilitates fusion events involving late endosomes and lysosomes. It ensures the fast and accurate assembly of trans -SNARE complexes to drive efficient membrane fusion.
A family of proteins with 4—12 repeats of a β-stranded blade that function as structural scaffolds for ligand binding, enzymatic activity and assembly of multiple protein complexes.
A protein domain that binds phosphatidylinositol lipids such as phosphatidylinositol 4,5-bisphosphate and proteins with high affinity and specificity. It functions in vesicle trafficking, cellular signalling and cytoskeletal remodelling. Intracellular movement from the cell periphery towards the nucleus mediated by the dynein—dynactin complex.
Also known as centripetal movement or minus-end transport. Intracellular movement from the nucleus towards the cell periphery mediated by kinesin motors. Also known as centrifugal movement or plus-end transport.
A multisubunit complex that associates peripherally with the lysosomal membrane to regulate lysosomal positioning by recruiting ARL8. It comprises eight subunits: BLOS1, BLOS2, snapin, KXD1, myrlysin, lyspersin, diaskedin and MEF2BNB.
A small ADP - ribosylation factor - like RAS family GTPase that mediates kinesin-driven lysosome transport, and also regulates lysosome fusion by recruiting the HOPS complex. A group of inherited metabolic disorders characterized by abnormal storage of toxic materials.
They result from deficiencies of lysosomal enzymes or transporters. The Atg1 complex consists of the protein kinase Atg1 together with Atg13, Atg17, Atg31 and Atg Activation of this complex triggers the initiation of autophagy.
A complex, containing the phosphatidylinositol 3-phosphate PtdIns3P kinase Vps34 together with Vps15, Atg6 and Atg14, that generates PtdIns3P at the autophagosome formation site to recruit downstream effectors for autophagosome formation.
It is catalysed by O -GlcNAc transferase OGT and reversed by O -GlcNAcase OGA. Vesicles whose formation is driven by COPII coat proteins. COPII vesicles transport proteins from the endoplasmic reticulum to the Golgi apparatus. A protein that facilitates the GTPase activity of small GTPases resulting in hydrolysis of GTP to GDP and phosphate.
A response elicited by accumulation of unfolded proteins in the endoplasmic reticulum ER. It promotes several intracellular signal transduction pathways, collectively known as the unfolded protein response, to restore ER homeostasis.
A process triggered by multivalent interactions of constituents, resulting in their concentration in a confined liquid-like compartment that stably coexists with the surrounding liquid milieu. A multiprotein complex that interacts with transcription factors and RNA polymerase II and functions as a transcriptional co-activator.
An aggregation-prone protein that accumulates in Lewy bodies and plays a central role in the pathogenesis of Parkinson disease and other synucleinopathies. An expansion of the CAG-trinucleotide repeat in disease-related genes resulting in polyglutamine expansion disorders such as Huntington disease and spinocerebellar ataxias.
An X-linked, recessive lower motor neuron disease caused by a CAG polyglutamine repeat expansion within the first exon of the androgen receptor gene. A mixture of small peptides 37—43 amino acids derived from sequential secretase-mediated proteolytic processing of amyloid precursor protein APP.
Aβ aggregates into insoluble neurotoxic plaques in patients with Alzheimer disease. A major component of the neurofibrillary tangles found in a number of neurodegenerative diseases, including Alzheimer disease.
A multisystem disorder agenesis of the corpus callosum, progressive microcephaly, cataracts, hypopigmentation, combined immunodeficiency and recurrent infections, and cardiomyopathy caused by mutations in the autophagy gene EPG5. A disease caused by WDR45 mutations.
Patients show static psychomotor retardation in early childhood, then develop sudden-onset progressive parkinsonism, dystonia and cognitive impairment in early adulthood.
The receptor for chaperone-mediated autophagy. Mutations in the gene encoding LAMP2 are causatively linked with Danon disease. Endoplasmic reticulum-derived vesicles that tune endoplasmic reticulum-associated degradation activity by targeting regulators of endoplasmic reticulum-associated degradation such as EDEM1 to lysosomes for degradation.
A process involving extraction of misfolded proteins from the endoplasmic reticulum ER for subsequent proteasomal degradation. Apparatuses used by bacteria to secrete virulence factors from the bacterial cytosol into host cells or the host cell environment.
A cell-penetrating peptide that induces autophagy. Eighteen amino acids from the evolutionarily conserved domain of beclin 1 are covalently coupled with the HIV-1 Tat protein transduction domain. Substances that are taken up selectively into lysosomes irrespective of their chemical nature or uptake mechanism.
Selenium CSS selectors reconstitute Autophagy machinery early events in autophagy i Autoophagy vitro. Autophagy machinery of an Autpohagy vesicle from experimental data and bioinformatics predictions. The interaction machlnery Autophagy machinery Atg Autophagy machinery machunery their Autophagy machinery enables Atg8 to be anchored in machonery membrane - an mahcinery step for the development of the phagophore. Even in cells savings are made: Autophagy is a natural regulatory mechanism of the cell, which ensures that unnecessary or defective components are dismantled recycled. In autophagy, cytoplasmic components like mitochondria are engulfed by a growing double membrane termed phagophore. Upon closure, this structure now called autophagosome isolates its cargo from the rest of the cell within a double-membraned vesicle. It fuses with a lysosome to start the process of waste management and disposal.
Seiner erreichte noch nicht.
Ich entschuldige mich, aber meiner Meinung nach sind Sie nicht recht. Es ich kann beweisen. Schreiben Sie mir in PM, wir werden umgehen.